Hur man använder regular expressions i R
Dealing with Regular Expressions
A regular expression (aka regex) is a sequence of characters that define a search pattern, mainly for use in pattern matching with text strings. Typically, regex patterns consist of a combination of alphanumeric characters as well as special characters. The pattern can also be as simple as a single character or it can be more complex and include several characters.
To understand how to work with regular expressions in R, we need to consider two primary features of regular expressions. One has to do with the syntax, or the way regex patterns are expressed in R. The other has to do with the functionsused for regex matching in R. In this section, we will cover both of these aspects. First, I cover the syntax that allow you to perform pattern matching functions with meta characters, character and POSIX classes, and quantifiers. This will provide you with the basic understanding of the syntax required to establish the pattern to find. Then I cover the functions provided in base R and in the stringr package you can apply to identify, extract, replace, and split parts of character strings based on the regex pattern specified.
Regex Syntax
At first glance (and second, third,…) the regex syntax can appear quite confusing. This section will provide you with the basic foundation of regex syntax; however, realize that there is a plethora of resources available that will give you far more detailed, and advanced, knowledge of regex syntax. To read more about the specifications and technicalities of regex in R you can find help at help(regex) or help(regexp).
Metacharacters
Metacharacters consist of non-alphanumeric symbols such as:. \\\ | ( ) [ { $ * + ?
To match metacharacters in R you need to escape them with a double backslash “\\”. The following displays the general escape syntax for the most common metacharacters:
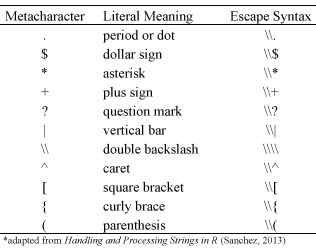
The following provides examples to show how to use the escape syntax to find and replace metacharacters. For information on the sub and gsub functions used in this example visit the main regex functions section.
# substitute $ with !
sub(pattern = "\\$", "\\!", "I love R$")
## [1] "I love R!"
# substitute ^ with carrot
sub(pattern = "\\^", "carrot", "My daughter has a ^ with almost every meal!")
## [1] "My daughter has a carrot with almost every meal!"
# substitute \\ with whitespace
gsub(pattern = "\\\\", " ", "I\\need\\space")
## [1] "I need space"
Sequences
To match a sequence of characters we can apply short-hand notation which captures the fundamental types of sequences. The following displays the general syntax for these common sequences:

The following provides examples to show how to use the anchor syntax to find and replace sequences. For information on the gsub function used in this example visit the main regex functions section.
# substitute any digit with an underscore
gsub(pattern = "\\d", "_", "I'm working in RStudio v.0.99.484")
## [1] "I'm working in RStudio v._.__.___"
# substitute any non-digit with an underscore
gsub(pattern = "\\D", "_", "I'm working in RStudio v.0.99.484")
## [1] "_________________________0_99_484"
# substitute any whitespace with underscore
gsub(pattern = "\\s", "_", "I'm working in RStudio v.0.99.484")
## [1] "I'm_working_in_RStudio_v.0.99.484"
# substitute any wording with underscore
gsub(pattern = "\\w", "_", "I'm working in RStudio v.0.99.484")
## [1] "_'_ _______ __ _______ _._.__.___"
Character classes
To match one of several characters in a specified set we can enclose the characters of concern with square brackets [ ]. In addition, to match any characters not in a specified character set we can include the caret ^ at the beginning of the set within the brackets. The following displays the general syntax for common character classes but these can be altered easily as shown in the examples that follow:

The following provides examples to show how to use the anchor syntax to match character classes. For information on the grep function used in this example visit the main regex functions section.
x <- c("RStudio", "v.0.99.484", "2015", "09-22-2015", "grep vs. grepl")
# find any strings with numeric values between 0-9
grep(pattern = "[0-9]", x, value = TRUE)
## [1] "v.0.99.484" "2015" "09-22-2015"
# find any strings with numeric values between 6-9
grep(pattern = "[6-9]", x, value = TRUE)
## [1] "v.0.99.484" "09-22-2015"
# find any strings with the character R or r
grep(pattern = "[Rr]", x, value = TRUE)
## [1] "RStudio" "grep vs. grepl"
# find any strings that have non-alphanumeric characters
grep(pattern = "[^0-9a-zA-Z]", x, value = TRUE)
## [1] "v.0.99.484" "09-22-2015" "grep vs. grepl"
POSIX character classes
Closely related to regex character classes are POSIX character classes which are expressed in double brackets [[ ]].

The following provides examples to show how to use the anchor syntax to match POSIX character classes. For information on the grep function used in this example visit the main regex functions section.
x <- "I like beer! #beer, @wheres_my_beer, I like R (v3.2.2) #rrrrrrr2015"
# remove space or tabs
gsub(pattern = "[[:blank:]]", replacement = "", x)
## [1] "Ilikebeer!#beer,@wheres_my_beer,IlikeR(v3.2.2)#rrrrrrr2015"
# replace punctuation with whitespace
gsub(pattern = "[[:punct:]]", replacement = " ", x)
## [1] "I like beer beer wheres my beer I like R v3 2 2 rrrrrrr2015"
# remove alphanumeric characters
gsub(pattern = "[[:alnum:]]", replacement = "", x)
## [1] " ! #, @__, (..) #"
Quantifiers
When we want to match a certain number of characters that meet a certain criteria we can apply quantifiers to our pattern searches. The quantifiers we can use are:

The following provides examples to show how to use the quantifier syntax to match a certain number of characters patterns. For information on the grepfunction used in this example visit the main regex functions section. Note that state.name is a built in dataset within R that contains all the U.S. state names.
# match states that contain z
grep(pattern = "z+", state.name, value = TRUE)
## [1] "Arizona"
# match states with two s
grep(pattern = "s{2}", state.name, value = TRUE)
## [1] "Massachusetts" "Mississippi" "Missouri" "Tennessee"
# match states with one or two s
grep(pattern = "s{1,2}", state.name, value = TRUE)
## [1] "Alaska" "Arkansas" "Illinois" "Kansas"
## [5] "Louisiana" "Massachusetts" "Minnesota" "Mississippi"
## [9] "Missouri" "Nebraska" "New Hampshire" "New Jersey"
## [13] "Pennsylvania" "Rhode Island" "Tennessee" "Texas"
## [17] "Washington" "West Virginia" "Wisconsin"
Regex Functions in Base R
R contains a set of functions in the base package that we can use to find pattern matches. Alternatively, the R package stringr also provides several functions for regex operations. This section covers the base R functions that provide pattern finding, pattern replacement, and string splitting capabilities.
Pattern Finding Functions
There are five functions that provide pattern matching capabilities. The three functions that I provide examples for are ones that are most common. The two other functions which I do not illustrate are gregexpr() and regexec()which provide similar capabilities as regexpr() but with the output in list form.
- Pattern matching with values or indices as outputs
- Pattern matching with logical (TRUE/FALSE) outputs
- Identifying the location in the string where the patter exists
grep( )
To find a pattern in a character vector and to have the element values or indices as the output use grep():
# use the built in data set `state.division`
head(as.character(state.division))
## [1] "East South Central" "Pacific" "Mountain"
## [4] "West South Central" "Pacific" "Mountain"
# find the elements which match the patter
grep("North", state.division)
## [1] 13 14 15 16 22 23 25 27 34 35 41 49
# use 'value = TRUE' to show the element value
grep("North", state.division, value = TRUE)
## [1] "East North Central" "East North Central" "West North Central"
## [4] "West North Central" "East North Central" "West North Central"
## [7] "West North Central" "West North Central" "West North Central"
## [10] "East North Central" "West North Central" "East North Central"
# can use the 'invert' argument to show the non-matching elements
grep("North | South", state.division, invert = TRUE)
## [1] 2 3 5 6 7 8 9 10 11 12 19 20 21 26 28 29 30 31 32 33 37 38 39
## [24] 40 44 45 46 47 48 50
grepl( )
To find a pattern in a character vector and to have logical (TRUE/FALSE) outputs use grep():
grepl("North | South", state.division)
## [1] TRUE FALSE FALSE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [12] FALSE TRUE TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE TRUE
## [23] TRUE TRUE TRUE FALSE TRUE FALSE FALSE FALSE FALSE FALSE FALSE
## [34] TRUE TRUE TRUE FALSE FALSE FALSE FALSE TRUE TRUE TRUE FALSE
## [45] FALSE FALSE FALSE FALSE TRUE FALSE
# wrap in sum() to get the count of matches
sum(grepl("North | South", state.division))
## [1] 20
regexpr( )
To find exactly where the pattern exists in a string use regexpr():
x <- c("v.111", "0v.11", "00v.1", "000v.", "00000")
regexpr("v.", x)
## [1] 1 2 3 4 -1
## attr(,"match.length")
## [1] 2 2 2 2 -1
## attr(,"useBytes")
## [1] TRUE
The output of regexpr() can be interepreted as follows. The first element provides the starting position of the match in each element. Note that the value -1 means there is no match. The second element (attribute “match length”) provides the length of the match. The third element (attribute “useBytes”) has a value TRUE meaning matching was done byte-by-byte rather than character-by-character.
Pattern Replacement Functions
In addition to finding patterns in character vectors, its also common to want to replace a pattern in a string with a new patter. There are two options for this:
sub( )
To replace the first matching occurrence of a pattern use sub():
new <- c("New York", "new new York", "New New New York")
new
## [1] "New York" "new new York" "New New New York"
# Default is case sensitive
sub("New", replacement = "Old", new)
## [1] "Old York" "new new York" "Old New New York"
# use 'ignore.case = TRUE' to perform the obvious
sub("New", replacement = "Old", new, ignore.case = TRUE)
## [1] "Old York" "Old new York" "Old New New York"
gsub( )
To replace all matching occurrences of a pattern use gsub():
# Default is case sensitive
gsub("New", replacement = "Old", new)
## [1] "Old York" "new new York" "Old Old Old York"
# use 'ignore.case = TRUE' to perform the obvious
gsub("New", replacement = "Old", new, ignore.case = TRUE)
## [1] "Old York" "Old Old York" "Old Old Old York"
Splitting Character Vectors
To split the elements of a character string use strsplit():
x <- paste(state.name[1:10], collapse = " ")
# output will be a list
strsplit(x, " ")
## [[1]]
## [1] "Alabama" "Alaska" "Arizona" "Arkansas" "California"
## [6] "Colorado" "Connecticut" "Delaware" "Florida" "Georgia"
# output as a vector rather than a list
unlist(strsplit(x, " "))
## [1] "Alabama" "Alaska" "Arizona" "Arkansas" "California"
## [6] "Colorado" "Connecticut" "Delaware" "Florida" "Georgia"
Regex Functions with stringr
Similar to basic string manipulation, the stringr package also offers regex functionality. In some cases the stringr performs the same functions as certain base R functions but with more consistent syntax. In other cases stringr offers additional functionality that is not available in the base R functions. The stringr functions we’ll cover focus on detecting, locating, extracting, and replacing patterns along with string splitting.
# install stringr package
install.packages("stringr")
# load package
library(stringr)
Detecting Patterns
To detect whether a pattern is present (or absent) in a string vector use the str_detect(). This function is a wrapper for grepl().
# use the built in data set 'state.name'
head(state.name)
## [1] "Alabama" "Alaska" "Arizona" "Arkansas" "California"
## [6] "Colorado"
str_detect(state.name, pattern = "New")
## [1] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [12] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [23] FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE FALSE
## [34] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [45] FALSE FALSE FALSE FALSE FALSE FALSE
# count the total matches by wrapping with sum
sum(str_detect(state.name, pattern = "New"))
## [1] 4
Locating Patterns
To locate the occurrences of patterns stringr offers two options: i) locate the first matching occurrence or ii) locate all occurrences. To locate the position of the first occurrence of a pattern in a string vector use str_locate(). The output provides the starting and ending position of the first match found within each element.
x <- c("abcd", "a22bc1d", "ab3453cd46", "a1bc44d")
# locate 1st sequence of 1 or more consecutive numbers
str_locate(x, "[0-9]+")
## start end
## [1,] NA NA
## [2,] 2 3
## [3,] 3 6
## [4,] 2 2
To locate the positions of all pattern match occurrences in a character vector use str_locate_all(). The output provides a list the same length as the number of elements in the vector. Each list item will provide the starting and ending positions for each pattern match occurrence in its respective element.
# locate all sequences of 1 or more consecutive numbers
str_locate_all(x, "[0-9]+")
## [[1]]
## start end
##
## [[2]]
## start end
## [1,] 2 3
## [2,] 6 6
##
## [[3]]
## start end
## [1,] 3 6
## [2,] 9 10
##
## [[4]]
## start end
## [1,] 2 2
## [2,] 5 6
Extracting Patterns
For extracting a string containing a pattern, stringr offers two primary options: i) extract the first matching occurrence or ii) extract all occurrences. To extract the first occurrence of a pattern in a character vector use str_extract(). The output will be the same length as the string and if no match is found the output will be NA for that element.
y <- c("I use R #useR2014", "I use R and love R #useR2015", "Beer")
str_extract(y, pattern = "R")
## [1] "R" "R" NA
To extract all occurrences of a pattern in a character vector use str_extract_all(). The output provides a list the same length as the number of elements in the vector. Each list item will provide the matching pattern occurrence within that relative vector element.
str_extract_all(y, pattern = "[[:punct:]]*[a-zA-Z0-9]*R[a-zA-Z0-9]*")
## [[1]]
## [1] "R" "#useR2014"
##
## [[2]]
## [1] "R" "R" "#useR2015"
##
## [[3]]
## character(0)
Replacing Patterns
For extracting a string containing a pattern, stringr offers two options: i) replace the first matching occurrence or ii) replace all occurrences. To replace the first occurrence of a pattern in a character vector use str_replace(). This function is a wrapper for sub().
cities <- c("New York", "new new York", "New New New York")
cities
## [1] "New York" "new new York" "New New New York"
# case sensitive
str_replace(cities, pattern = "New", replacement = "Old")
## [1] "Old York" "new new York" "Old New New York"
# to deal with case sensitivities use Regex syntax in the 'pattern' argument
str_replace(cities, pattern = "[N]*[n]*ew", replacement = "Old")
## [1] "Old York" "Old new York" "Old New New York"
To extract all occurrences of a pattern in a character vector use str_replace_all(). This function is a wrapper for gsub().
str_replace_all(cities, pattern = "[N]*[n]*ew", replacement = "Old")
## [1] "Old York" "Old Old York" "Old Old Old York"
String Splitting
To split the elements of a character string use str_split(). This function is a wrapper for strsplit().
z <- "The day after I will take a break and drink a beer."
str_split(z, pattern = " ")
## [[1]]
## [1] "The" "day" "after" "I" "will" "take" "a" "break"
## [9] "and" "drink" "a" "beer."
a <- "Alabama-Alaska-Arizona-Arkansas-California"
str_split(a, pattern = "-")
## [[1]]
## [1] "Alabama" "Alaska" "Arizona" "Arkansas" "California"
Note that the output of strs_plit() is a list. To convert the output to a simple atomic vector simply wrap in unlist():
unlist(str_split(a, pattern = "-"))
## [1] "Alabama" "Alaska" "Arizona" "Arkansas" "California"
Additional Resources
Character string data are often considered semi-structured data. Text can be structured in a specified field; however, the quality and consistency of the text input can be far from structured. Consequently, managing and manipulating character strings can be extremely tedious and unique to each data wrangling process. As a result, taking the time to learn the nuances of dealing with character strings and regex functions can provide a great return on investment; however, the functions and techniques required will likey be greater than what I could offer here. So here are additional resources that are worth reading and learning from: